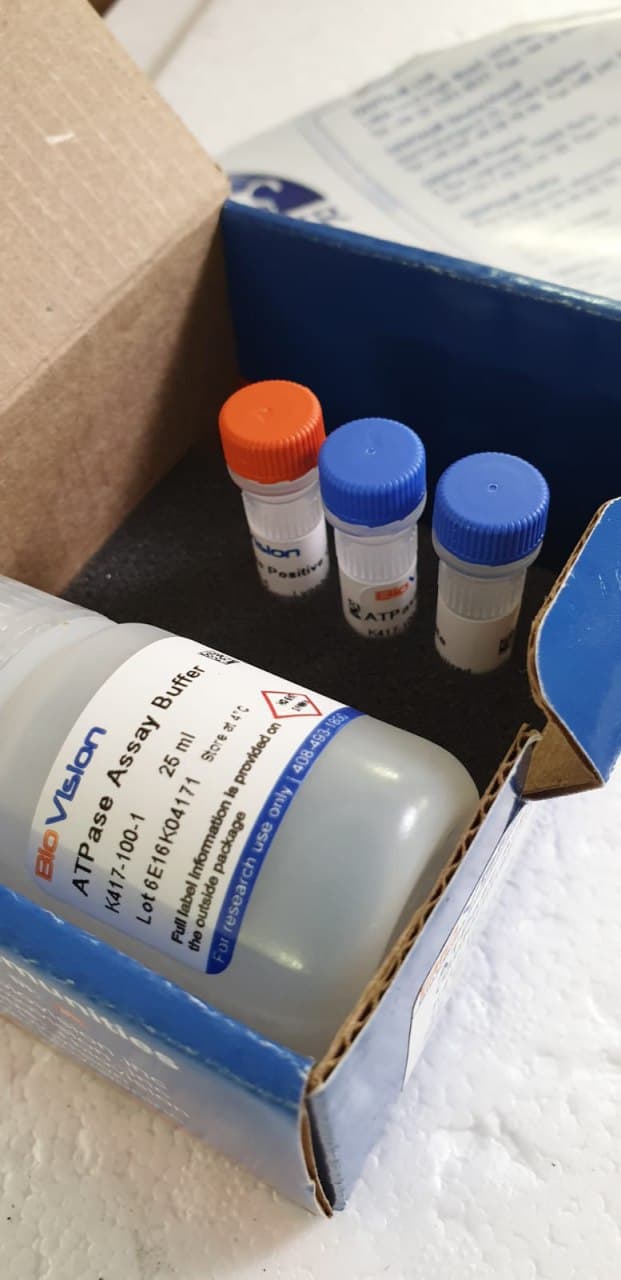
The wide selection of protein constructions and capabilities outcomes from the various properties of the 20 canonical amino acids. The usually accepted speculation is that early protein evolution was related to enrichment of a primordial alphabet, thereby enabling elevated protein catalytic efficiencies and useful diversification. Aromatic amino acids have been possible among the many final additions to genetic code. The foremost goal of this research was to take a look at whether or not enzyme catalysis can happen with out the aromatic residues (aromatics) by finding out the construction and performance of dephospho-CoA kinase (DPCK) following aromatic residue depletion. We designed two variants of a putative DPCK from Aquifex aeolicus by substituting (i) Tyr, Phe and Trp or (ii) all aromatics (together with His).
Their structural characterization signifies that substituting the aromatics doesn’t markedly alter their secondary constructions however does considerably loosen their facet chain packing and improve their sizes. Both variants nonetheless possess ATPase exercise, though with 150-300 occasions decrease effectivity compared with the wild-type phosphotransferase exercise. The switch of the phosphate group to the dephospho-CoA substrate turns into closely uncoupled and solely the His-containing variant remains to be in a position to carry out the phosphotransferase response.
These information assist the speculation that proteins within the early phases of life may assist catalytic actions, albeit with low efficiencies. An noticed important contraction upon ligand binding is probably going essential for applicable group of the lively website. Formation of agency hydrophobic cores, which allow the meeting of stably structured lively websites, is usually recommended to present a selective benefit for including the aromatic residues. This article is protected by copyright. All rights reserved.
Transcriptional response regulators (TRR) are essentially the most considerable sign transducers in prokaryotic methods that mediate intracellular modifications in response to environmental indicators. They are concerned in a wide selection of organic processes that enable micro organism to persist particularly habitats. There is robust proof that the bacterial habitat and their way of life affect the dimensions of their TRR genetic repertoire. Therefore, it might be anticipated that the evolution of bacterial genomes might be linked to pure choice processes. To take a look at this speculation, we explored the evolutionary dynamics of TRR genes of the extensively studied Harveyi clade of the genus Vibrio on the molecular and genomic ranges.
Our outcomes counsel that the TRR genetic repertoire of the species belonging to the Harveyi clade is a product of genomic discount and enlargement. The gene loss and features that drive their genomic discount and enlargement might be attributed to pure choice and random genetic drift. It appears that pure choice acts to preserve the ancestral state of core TRR genes (shared by all species) by purifying processes and might be driving the loss of some accent (present in sure species) genes by means of the diversification of sequences. The neutrality noticed in gene achieve might be attributed to spontaneous occasions as horizontal gene switch pushed by stochastic occasions as happens in random genetic drift.
Transposable components (TEs) are ubiquitous cell genetic components that maintain each disruptive and adaptive potential for species. It has lengthy been postulated that their exercise could also be triggered by hybridization, a speculation that obtained combined assist from research in numerous species. While host protection mechanisms in opposition to TEs are being elucidated, the growing quantity of genomic information and bioinformatic instruments specialised in TE detection allow in-depth characterization of TEs on the ranges of species and populations. Here, I borrow components from the genome ecology concept to illustrate how data of the range of TEs and host protection mechanisms could assist predict the exercise of TEs within the face of hybridization, and the way present limitations make this activity particularly difficult
